Advanced Microscopy
Scientists involved: Michael Bachmann, Prof. Dr. Martin Bastmeyer
Funding: Center for Functional Nanostructures (CFN), Karlsruhe School of Optics and Photonics (KSOP)
Superresolution Microscopy
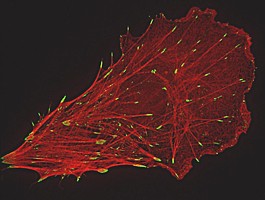
During the past decade, several light microscopic methods with resolutions beyond the diffraction limit have been developed. Among these approaches are structured illumination microscopy (SIM), photoactivated localization microscopy (PAL-M), direct stochastic optical reconstruction microscopy (dSTORM) and stimulated emission depletion STED. For SIM, a periodic pattern is projected into the specimen plane leading to moiré fringes and therefore improving resolution by capturing high-frequency information of the sample. SIM allows a resolution of 100 nm (xy) and 250 (z). PAL-M and dSTORM rely on the localization of single fluorescent molecules per time. To obtain spatially resolved images, cycles of illumination-detection-bleaching are required leading to acquisition times of 15-30 min. Here, resolution is only limited by statistical significance and values below 50 nm are possible. All methods have their pros and cons. A comparison would be interesting but is difficult to perform. Standard PALM and dSTORM require special fluorophores and buffer solutions in addition to total internal reflection illumination (TIRF) for z-axis discrimination, while SIM is usually performed in mounting medium but not limited to a narrow z-plane.
We are using a non-serial prototype of Elyra-PS.1 (by Carl Zeiss MicroImaging, Germany) combining both SIM and PALM/dSTORM and thus offering the possibility to compare these methods. To do so, cells were cultured on special high-index (HI, n = 1.66) fibronectin-coated cover slips. Cells were fixed with 4% paraformaldehyde and treated with monoclonal antibodies (against tubulin, Paxillin, or membrane-proteins). Cells were incubated with Alexa488-coupled secondary antibodies and Alexa546-coupled Phalloidin to label actin filaments and embedded in Mowiol. Culturing on HI-cover slips allows TIRF microscopy with a penetration depth of ≈ 100 nm in Mowiol by using a 100x/1.57 NA Zeiss objective. Since dSTORM requires reducing agents, β-mercaptoethylamine (MEA) was added to Mowiol at a concentration of 200 mM. This treatment leads to sufficient blinking of Alexa488 for dSTORM-imaging. In addition, these fixed samples also allows for relocating the same cells at a confocal laser scanning microscopy (CLSM).
Our comparisons reveal that SIM results in doubling the resolution of Paxillin-labeled focal contacts in comparison to CLSM. Within the same focal contacts, dSTORM leads to a further increase in resolution showing distinct labeled structures of 50 nm diameter. In contrast, the dense meshwork of actin filaments in lamellipodia is better resolved with SIM as compared to CLSM and dSTORM. Due to the punctate nature of the single molecule localization signal, dSTORM data do not show these actin networks as filamentous structures.
Optical Tweezers
Scientists involved: M. Bastmeyer
Since their invention in 1986 by A. Ashkin, optical tweezers have become increasingly valuable tools for research in the biological and physical sciences. Using a focused beam of light to trap and move matter, optical tweezers offer convenient, non-invasive access to processes at the mesoscopic scale. In this project, we use an experimental design which is based on an acousto-optical deflection system (AOD) which allows the steering and intensity modulation of an incoming laser beam.
In collaboration with the group of Prof. Clemens Bechinger (http://www.pi2.uni-stuttgart.de/Uebersicht.html) at the University Stuttgart we have build a setup consisting of a multiple optical tweezers controlled by an AOD. With this approach we can overcome the limitations of existing optical tweezers, namely the limited size of trapped particles and the small numbers of particles that can be handled by one tweezers. Laser tweezers offer the opportunity to measure very small forces in the sub-piconewton and piconewton regime. In our setup we can simultaneously trap a number of micron-sized polystyrene beads, manipulate them in a clearly defined manner and use them as handles to living cells.
The system is capable to independently trap several hundred submicron-sized particles with nanometer resolution within a lateral addressable range in the sample plane of 150x150 µm². Incorporated into an inverted custom-built microscope, the system is applied to manipulate cells and to measure cell adhesion forces using functionalized polystyrol beads.
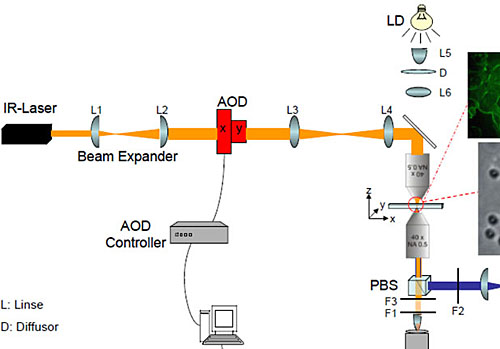
Fig. 1: Schematic drawing of the laser tweezer setup. (AOD: Acousto-Optical Deflector)
The setup is composed of an infrared Laser (Compass1064, Coherent), an acousto-optical deflector (AOD) with control unit (AA Optoelectronic and Aresis) and a custom-built microscope. To focus the laser beam an objective with a high numerical aperture is used (Plan-Neofluar 63x, 1.2 NA Korr, water immersion; Zeiss). For the second objective, which is used for signal detection, magnifications from 10x to 63x are available. A white light emitting diode (LED) serves as microscope illumination. For epifluorescence illumination an additional port in the lower part of the microscope is accessible and equipped with a high power lamp (X-Cite 120, Exfo).
To generate an optical trap the laser light is first expanded and then send through the AOD. The deflector allows both, beam steering and the generation of multiple, time shared traps with a switching rate of up to 50 kHz. Thus, several hundred submicron-sized particles can be trapped independently with nanometer resolution within a lateral addressable range in the sample plane of 150x150 µm². Behind the AOD the beam is led through a set of lenses and into the microscope where the beam is focused with a high numerical aperture objective. A second objective passes the signal on to a CCD camera, where it is recorded.
For biological experiments with living cells, a live cell imaging chamber is included in the sample plan. This chamber is temperature controlled (37°C) for the culturing of mammalian cell lines and is equipped with micro fluidic devices for the exchange of cell culture media.