Cell Mechanics
Scientists involved: Prof. Dr. Martin Bastmeyer, Dr. Kai Weißenbruch, Magdalena Fladung, Lucie Mißbach
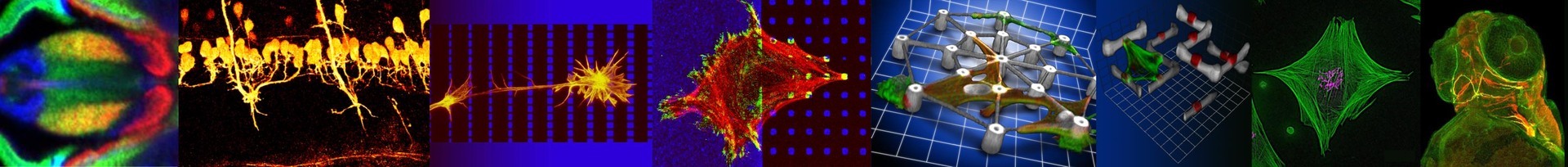
Intracellular forces generated by the actomyosin system act in fundamental cellular processes including morphogenesis, cell migration, cell adhesion and cytokinesis. Class II non-muscle myosins (NM II) are the primary contractile proteins in eukaryotic nonmuscle cells.
Mammalian cells express up to three different isoforms of NM II which differ in their biochemical features but largely overlap in spatial and temporal activity by assembling into homotypic as well as heterotypic bipolar minifilaments. Moreover, structure and molecular composition of the actomyosin system are highly dynamic and undergo a permanent remodelling. Therefore, a detailed understanding of how contractile units are coordinated on a subcellular scale is still missing. We are investigating how the actomyosin system emerges, how it is organized and how it ultimately guides processes like morphogenesis and cell migration.
Mechanoresponsive stress reactions of non-muscle cells
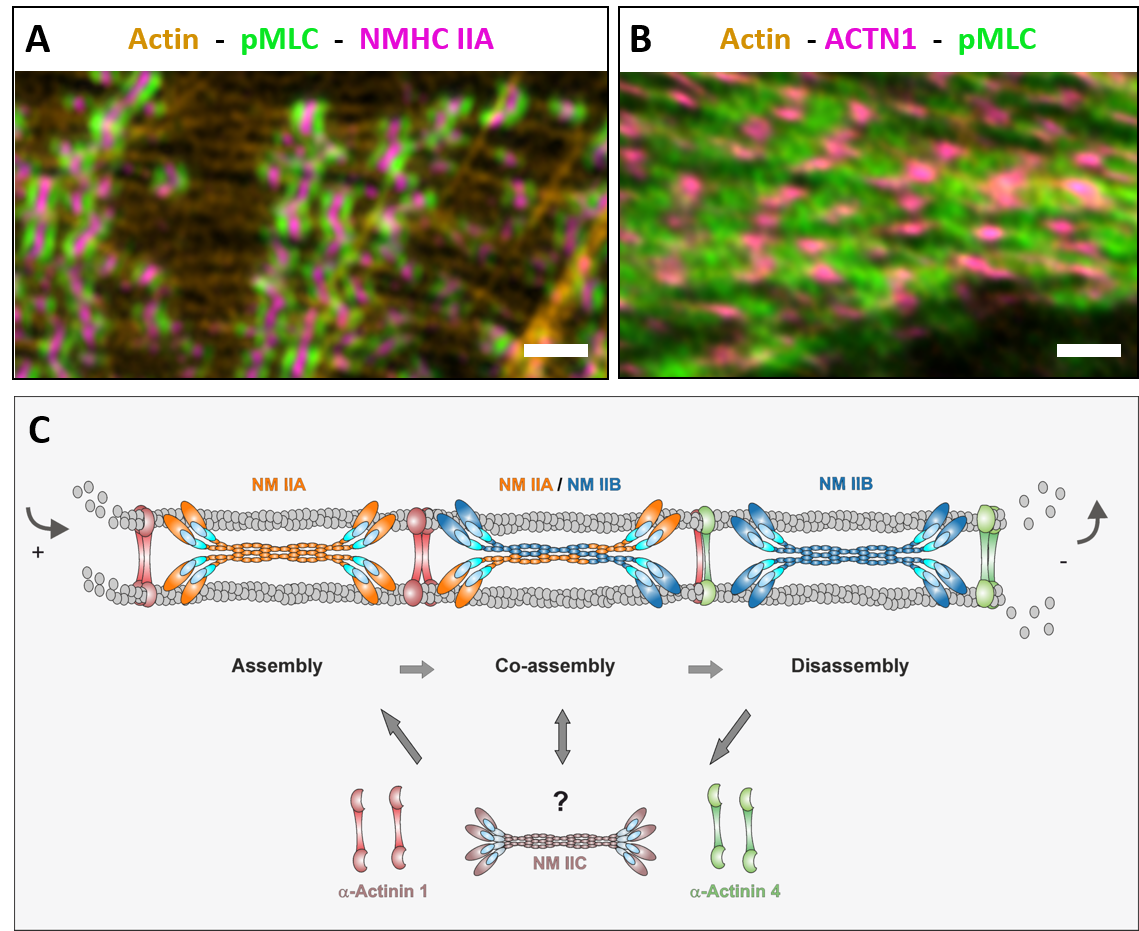
Manipulating single cells is primarily important to understand how they behave at higher-complexity levels, i.e. in tissues and organs. A challenging step towards the realization of controlled systems to study single cells in three dimensions is the implementation of techniques allowing for the precise positioning of distinct cell types in a 3D microenvironment.
By using polymeric materials with different affinity to proteins of the extracellular matrix (ECM), it is possible to exploit 2PL to realize 3D scaffolds with defined geometries and cell-adhesive and cell-repulsive regions (figs. A-B). This allows to carefully control cell shape and functions, as well as to investigate different behaviours (e.g. direction and intensity of exerted forces, activation of selected pathways).
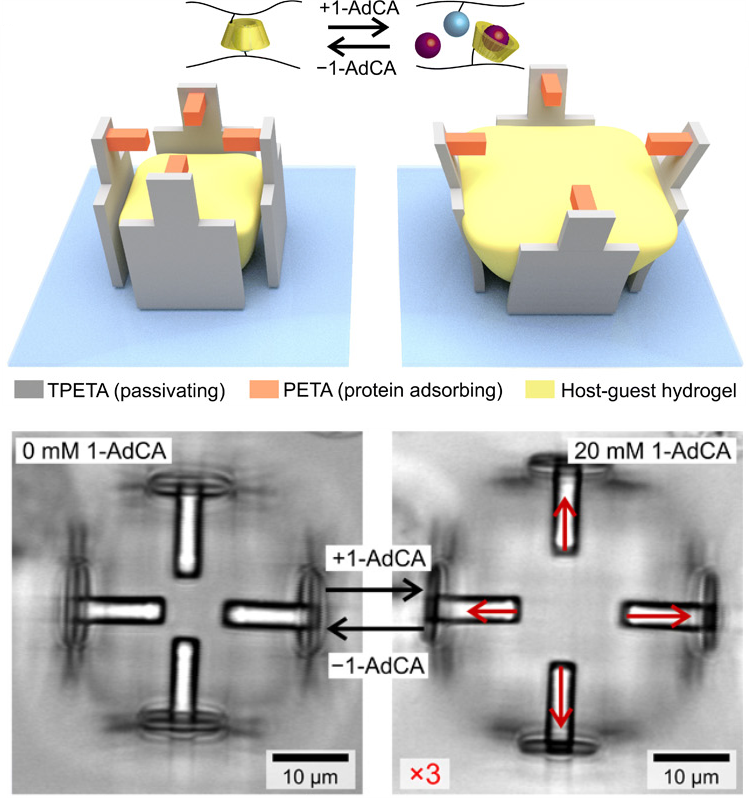
We developed a method to apply mechanical stretch-release cycles to single cells. Using phase contrast and 3D AiryScan fluorescence imaging, we monitor the cellular response dynamics during tensional homeostasis with both, high spatial and temporal resolution. Collecting this information allows us to i) extract the cellular forces exerted by the cells and ii) monitor the dynamic rearrangement of the contractile units with subcellular resolution.
Modelling of cell shape and cell forces (in cooperation with Prof. Dr. Ulrich Schwarz, Heidelberg)
Experimental data collected on structured substrates were complemented by theoretical models about cell shape formation, cellular forces and molecular dynamics of NM II molecules. In cooperation with the group of Prof. Ulrich Schwarz (Institute for Theoretical Physics and BioQuant, University Heidelberg), methods and concepts from mechanics, stochastic dynamics and non-linear dynamics were applied to theoretically describe the experimental data. Our goal is to gain a systems level understanding of how individual contractile units are coordinated to generate forces that determine the cellular phenotype.
Further reading:
Link, R., Weißenbruch, K. et al. (2023). Cell Shape and Forces in Elastic and Structured Environments: From Single Cells to Organoids. Advanced Functional Materials, https://doi.org/10.1002/adfm.202302145.
Weißenbruch, K., et al. "Nonmuscle myosin IIA dynamically guides regulatory light chain phosphorylation and assembly of nonmuscle myosin IIB." European Jornal of Cell Biology (2022): 151213.
Weißenbruch, K., et al. "Distinct roles of nonmuscle myosin II isoforms for establishing tension and elasticity during cell morphodynamics." eLife (2021): 10:e7188